Compute correlations using the tidyverse
This small example aims to provide some use cases for the tidyr package. Let’s generate some example data first:
library(lubridate)
library(tibble)
library(dplyr)
library(tidyr)
library(ggplot2)
library(forcats)
library(purrr)
set.seed(1234)
sales <- tibble(date = ymd(rep(c(20180101, 20180102, 20180103), 3)),
product = rep(c("A", "B", "C"), each = 3),
sales = sample(1:20, size = 9, replace = T))
sales## # A tibble: 9 x 3
## date product sales
## <date> <chr> <int>
## 1 2018-01-01 A 3
## 2 2018-01-02 A 13
## 3 2018-01-03 A 13
## 4 2018-01-01 B 13
## 5 2018-01-02 B 18
## 6 2018-01-03 B 13
## 7 2018-01-01 C 1
## 8 2018-01-02 C 5
## 9 2018-01-03 C 14We want to compute the correlation of the sales from products A, B and C. The base R function cor() takes a matrix or data.frame and computes the correlation between all the column pairs. Thus, first we need to convert the data.frame sales, which is in long form, to wide form with one column per product.
cor_matrix <-
sales %>%
spread(key = product, value = sales) %>%
select(-date) %>%
cor()
cor_matrix## A B C
## A 1.0000000 0.5000000 0.7370435
## B 0.5000000 1.0000000 -0.2167775
## C 0.7370435 -0.2167775 1.0000000To manipulate the correlation matrix using tidyverse-related functions we need to convert back the previous matrix to a long data.frame:
cor_tidy <-
cor_matrix %>%
as.data.frame() %>%
rownames_to_column(var = "product1") %>%
gather(key = product2, value = corr, -product1)
cor_tidy## product1 product2 corr
## 1 A A 1.0000000
## 2 B A 0.5000000
## 3 C A 0.7370435
## 4 A B 0.5000000
## 5 B B 1.0000000
## 6 C B -0.2167775
## 7 A C 0.7370435
## 8 B C -0.2167775
## 9 C C 1.0000000Now we can plot the correlation matrix using ggplot2, for instance with a heatmap:
ggplot(cor_tidy, aes(x = product1, y = product2, fill = corr)) +
geom_tile() +
scale_fill_gradient2(limits = c(-1, 1))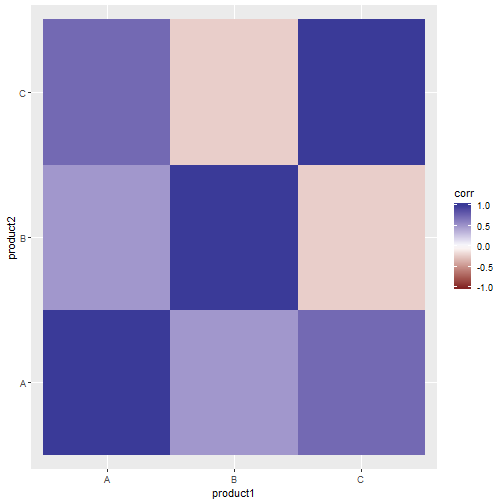
Another common way of representing correlation is a vertical barplot. For this type of plot we often want to ignore the diagonal and upper/lower triangle, and sort from lowest to highest:
cor_tidy %>%
filter(product1 != product2) %>%
distinct(products = paste(pmin(product1, product2),
pmax(product1, product2), sep = "_vs_"), .keep_all = TRUE) %>%
ggplot(aes(x = fct_reorder(products, corr), y = corr, fill = corr > 0.7)) +
geom_col(width = 0.7) +
coord_flip() +
ylim(-1, 1) +
xlab("products") +
theme(aspect.ratio = 1/3) +
scale_fill_discrete(name = "Cor > 0.7",
breaks = c(TRUE, FALSE),
labels = c("Yes", "No"))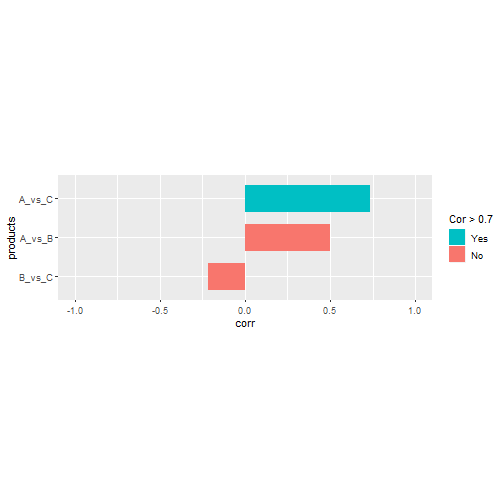
Here we are using a neat trick to ignore rows with duplicate product IDs ignoring its order (see this and this). The previous trick can be generalized to more than two columns, although it is not trivial (see this question for a base R solution). Let’s create first some example data:
values <- c("A", "B", "C")
df <- expand.grid(ID1 = values, ID2 = values, ID3 = values, stringsAsFactors = FALSE)
df## ID1 ID2 ID3
## 1 A A A
## 2 B A A
## 3 C A A
## 4 A B A
## 5 B B A
## 6 C B A
## 7 A C A
## 8 B C A
## 9 C C A
## 10 A A B
## 11 B A B
## 12 C A B
## 13 A B B
## 14 B B B
## 15 C B B
## 16 A C B
## 17 B C B
## 18 C C B
## 19 A A C
## 20 B A C
## 21 C A C
## 22 A B C
## 23 B B C
## 24 C B C
## 25 A C C
## 26 B C C
## 27 C C CWe would like to obtain unique ID combinations without taking order into account, that is, “AAB” and “ABA” are both the same:
distinct(df, ID = pmap_chr(select(df, starts_with("ID")),
~paste0(sort(c(...)), collapse="_")))## ID
## 1 A_A_A
## 2 A_A_B
## 3 A_A_C
## 4 A_B_B
## 5 A_B_C
## 6 A_C_C
## 7 B_B_B
## 8 B_B_C
## 9 B_C_C
## 10 C_C_CNote the c(...), since the .f argument in pmap() is a function with as many arguments as columns in the data frame (in contrast to base apply()). Thus we need to collect them all in a vector, which is then sorted and finally converted into a single value with paste(..., collapse="_").
